Additional links:
- Original class material
<< Previous lecture | Next lecture >>
todo clean
Probabilities in Machine Learning
In the ML lecture we said that a major assumption in Machine Learning is that there exists a (possibly stochastic) target function such that for any , and such that the datasets
are generated by considering independent identically distributed (i.i.d.) samples , where is the unknown distribution of the inputs, and considering for any . When is a stochastic function, we can consider the sampling process of as for any . In this setup, we can consider the decomposition
where is the joint distribution, is called prior distribution over , while is the likelihood or posterior distribution of given . With this framework, learning a Machine Learning model for any with parameters , can be reformulating as learning a parameterized distribution which maximizes the probability of observing , given .
Maximum Likelihood Estimation (MLE)
Intuitively, we would like to find parameters such that the probability of observing given is as high as possible. Consequently, we have to solve the optimization problem
Which is usually called Maximum Likelihood Estimation (MLE), because the parameters are chosen such that they maximize the likelihood . Since are independent under ,
and since is independent with for any , then
Consequently, \eqref{eq:mle_formulation1} becomes:
Since the logarithm function is monotonic, applying it to the optimization problem \eqref{eq:mle_formulation2} does not alterate its solution. Moreover, since for any function , , \eqref{eq:mle_formulation2} can be restated as
which is the classical formulation of an MLE problem. Note that in \eqref{eq:mle_formulation3}, the objective function has been decomposed into a sum over the datapoints for any , as a consequence of the being i.i.d.. This formulation is similar to what we required in the previous lecture, implying that we can use SGD to (approximately) solve \eqref{eq:mle_formulation3}.
Gaussian Assumption
To effectively solve \eqref{eq:mle_formulation3}, we must explicitely define . A common assumption, which is true for most of the scenarios, is to consider
where is a Gaussian distribution with mean and variance , with a parametric deterministic function of while is the variance of , which depends on the informations we have on the relationship between and (it will be clearer in the following example).
An interesting property of the Gaussian distribution is that if , then , where is a Standard Gaussian distribution.
To simplify the derivation below, assume that , so that and for any . It is known that if , then
thus
Consequently, MLE with Gaussian likelihood becomes
which can be reformulated as a Least Squares problem
where , while .
Polynomial Regression MLE
Now, consider a Regression model
and assume that
Then, by \eqref{eq:MLE_gaussian},
where
is the Vandermonde matrix associated with the vector and with feature vectors . Clearly, when , the regression model is a Polynomial Regression model and the associated Vandermonde matrix is the classical Vandermonde Matrix
Note that \eqref{eq:MLE_regression} defines a training procedure for a regression model. Indeed, it can be optimized by Gradient Descent (or its Stochastic variant), by solving
where
Direct solution by Normal Equations
Note that, since the learning problem is a Least Square problem of the form
then it can be solved directly by the Normal Equation method, i.e.
this solution can be compared with the convergence point of Gradient Descent, to check the differences.
MLE + Flexibility = Overfit
In polynomial regression, the most important parameter the user has to set is the degree of polynomial, . Indeed, when is low, the resulting model will be pretty rigid (not flexible), with the implication that it can potentially be unable to capture the complexity of the data. On the opposite side, if is too large, the resulting model is too flexible, and we end up learning the noise. The former situation, which is called underfitting, can be easily diagnoised by looking at a plot of the resulting model with respect to the data (or, equivalently, by checking the accuracy of the model). Conversely, when the model is too flexible, we are in an harder scenario known as overfitting. In overfitting, the model is not understanding the knowledge of the data, but it is memorizing the training set, usually resulting in optimal training error and bad test prediction.
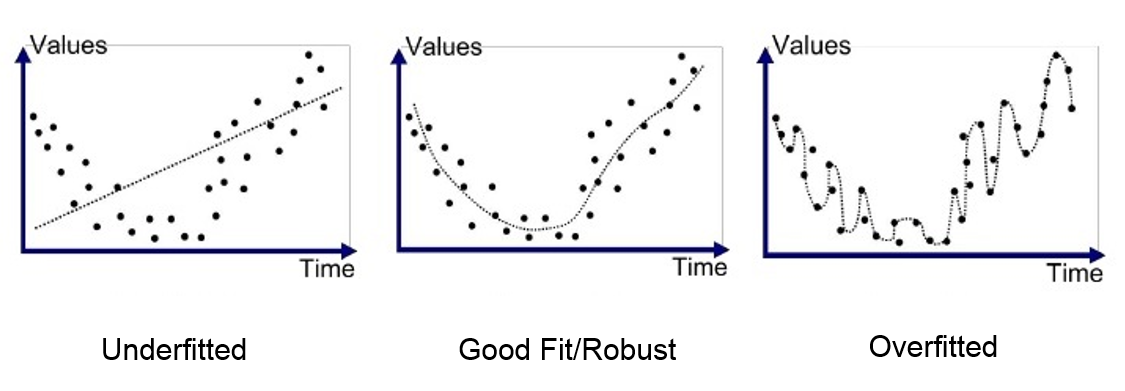
Ideally, when the data is generated by a noisy polynomial experiment, we would like to set as the real degree of such polynomial. Unfortunately, this is not always possible and indeed, spotting overfitting is the hardest issue to solve while working with Machine Learning.
Solving overfitting using the error plot
A common way to solve overfit, is to plot the error of the learnt model with respect to its complexity (i.e. the degree of the polynomial). In particular, for , one can train a polynomial regressor of degree over the training set and compute the training error as the average absolute error of the prediction on the training set, i.e.
and, for the same set of parameters, the test error
If we plot the training and test error with respect to the different values of , we will observe the following situation:
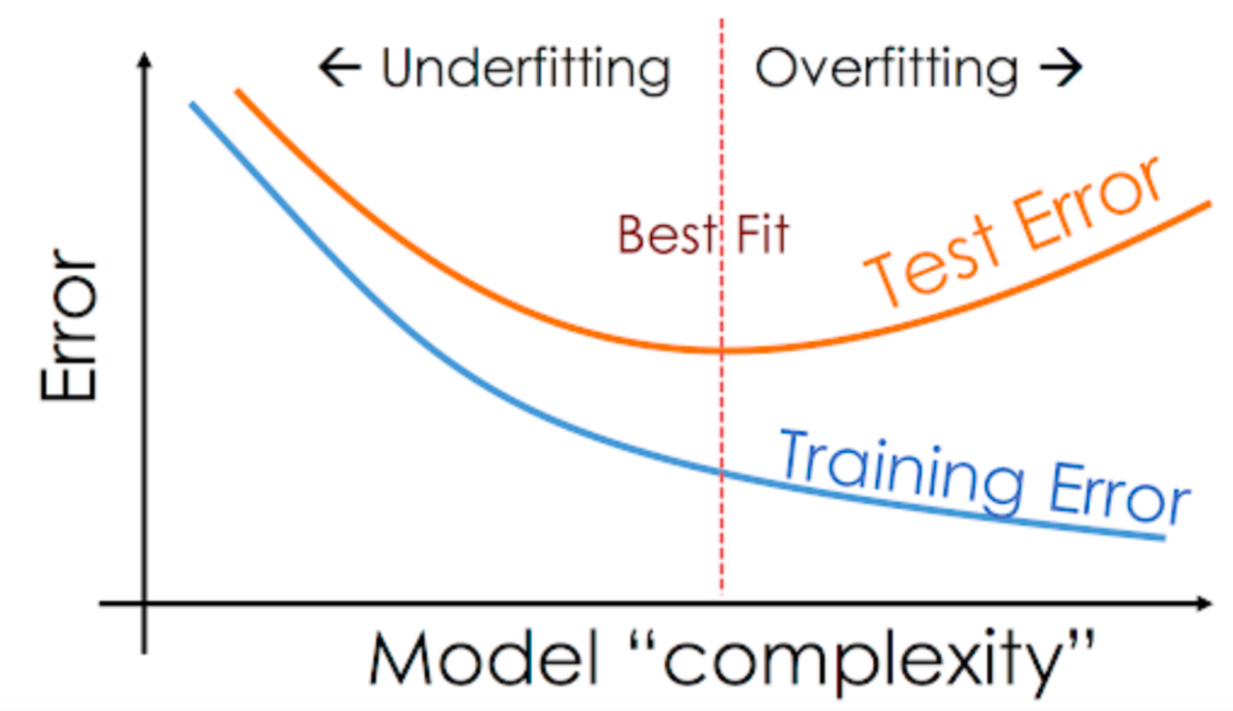
which will help us to find the correct parameter , not suffering underfitting nor overfitting.
A better solution: Maximum A Posteriori (MAP)
A completely different approach to overfitting is to change the perspective and stop using MLE. The idea is to reverse the problem and, instead of searching parameters such that the probability of observing the outcomes given the data is maximized, i.e. maximizing , as in MLE, try to maximize the probability that the observed data is , given the parameters . Mathematically, we are asked to solve the optimization problem
Since is called posterior distribution, this method is usually referred to as **Maximum A Posteriori (MAP).
Bayes Theorem
A problem of MAP, is that it is non-trivial to find a formulation for . Indeed, if with MLE the Gaussian assumption made sense, as a consequence of the hypothesis that the observations are obtained by corrupting a deterministic function of by Gaussian noise, this does not hold true for MAP, since in general the generation of given is not Gaussian.
Luckily, we can express the posterior distribution in terms of the likelihood (which we know to be Gaussian) and the prior , as a consequence of Bayes Theorem. Indeed, it holds
Gaussian assumption on MAP
For what we observed above, the posterior distribution can be rewritten as a function of the likelihood and the prior . Thus, \eqref{eq:MAP_formulation1} can be rewritten as
With the same trick we used in MLE, we can change it to a minimum point estimation by changing the sign of the function and by taking the logarithm. We obtain,
where we removed since it is constant in .
Since are i.i.d. by hypothesis and by following the same procedure of MLE, we can split \eqref{eq:MAP_formulation3} into a sum over datapoints, as
Now, if we assume that , the same computation we did in MLE implies
To complete the derivation, we have to rewrite in a meaningful way, to be able to perform the optimization. To do that, it is common to assume that , a Gaussian distribution with zero mean and variance . Under this assumption,
and consequently
where is a positive parameter, usually called regularization parameter. This equation is the final MAP loss function under Gaussian assumption for both and . Clearly, it is another Least Squares problem which can be solved by Gradient Descent or Stochastic Gradient Descent.
When is a polynomial regression model, , then
can be also solved by Normal Equations, as
Ridge Regression and LASSO
When the Gaussian assumption is used for both the likelihood and the prior , the resulting MAP is usually called Ridge Regression in the literature. On the contrary, if is Gaussian and is a Laplacian distribution with mean 0 and variance , then
and consequently (prove it by exercise)
the resulting model is called LASSO, and it is the basis for most of the classical, state-of-the-art regression models.